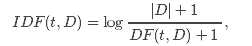
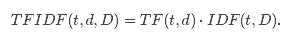
from pyspark import SparkContext from pyspark.mllib.feature import HashingTF sc = SparkContext() # Load documents (one per line). documents = sc.textFile("...").map(lambda line: line.split(" ")) hashingTF = HashingTF() tf = hashingTF.transform(documents) from pyspark.mllib.feature import IDF # ... continue from the previous example tf.cache()idf = IDF().fit(tf) tfidf = idf.transform(tf) # ... continue from the previous example tf.cache()idf = IDF(minDocFreq=2).fit(tf) tfidf = idf.transform(tf)
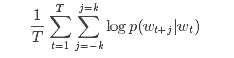
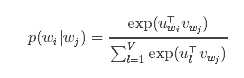
from pyspark import SparkContext from pyspark.mllib.feature import Word2Vec sc = SparkContext(appName=‘Word2Vec‘) inp = sc.textFile("text8_lines").map(lambda row: row.split(" ")) word2vec = Word2Vec() model = word2vec.fit(inp) synonyms = model.findSynonyms(‘china‘, 40) for word, cosine_distance in synonyms: print("{}: {}".format(word, cosine_distance))
from pyspark.mllib.util import MLUtils from pyspark.mllib.linalg import Vectors from pyspark.mllib.feature import StandardScaler data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") label = data.map(lambda x: x.label) features = data.map(lambda x: x.features) scaler1 = StandardScaler().fit(features) scaler2 = StandardScaler(withMean=True, withStd=True).fit(features) # scaler3 is an identical model to scaler2, and will produce identical transformations scaler3 = StandardScalerModel(scaler2.std, scaler2.mean) # data1 will be unit variance. data1 = label.zip(scaler1.transform(features)) # Without converting the features into dense vectors, transformation with zero mean will raise # exception on sparse vector. # data2 will be unit variance and zero mean. data2 = label.zip(scaler1.transform(features.map(lambda x: Vectors.dense(x.toArray()))))
from pyspark.mllib.util import MLUtils from pyspark.mllib.linalg import Vectors from pyspark.mllib.feature import Normalizer data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") labels = data.map(lambda x: x.label) features = data.map(lambda x: x.features) normalizer1 = Normalizer() normalizer2 = Normalizer(p=float("inf")) # Each sample in data1 will be normalized using $L^2$ norm. data1 = labels.zip(normalizer1.transform(features)) # Each sample in data2 will be normalized using $L^\infty$ norm. data2 = labels.zip(normalizer2.transform(features))
import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.feature.ChiSqSelector; import org.apache.spark.mllib.feature.ChiSqSelectorModel; import org.apache.spark.mllib.linalg.Vectors; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.util.MLUtils; SparkConf sparkConf = new SparkConf().setAppName("JavaChiSqSelector"); JavaSparkContext sc = new JavaSparkContext(sparkConf); JavaRDD<LabeledPoint> points = MLUtils.loadLibSVMFile(sc.sc(), "data/mllib/sample_libsvm_data.txt").toJavaRDD().cache(); // Discretize data in 16 equal bins since ChiSqSelector requires categorical features // Even though features are doubles, the ChiSqSelector treats each unique value as a category JavaRDD<LabeledPoint> discretizedData = points.map( new Function<LabeledPoint, LabeledPoint>() { @Override public LabeledPoint call(LabeledPoint lp) { final double[] discretizedFeatures = new double[lp.features().size()]; for (int i = 0; i < lp.features().size(); ++i) { discretizedFeatures[i] = Math.floor(lp.features().apply(i) / 16); } return new LabeledPoint(lp.label(), Vectors.dense(discretizedFeatures)); } }); // Create ChiSqSelector that will select top 50 of 692 features ChiSqSelector selector = new ChiSqSelector(50); // Create ChiSqSelector model (selecting features) final ChiSqSelectorModel transformer = selector.fit(discretizedData.rdd()); // Filter the top 50 features from each feature vector JavaRDD<LabeledPoint> filteredData = discretizedData.map( new Function<LabeledPoint, LabeledPoint>() { @Override public LabeledPoint call(LabeledPoint lp) { return new LabeledPoint(lp.label(), transformer.transform(lp.features())); } }); sc.stop();
import java.util.Arrays; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.mllib.feature.ElementwiseProduct; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; // Create some vector data; also works for sparse vectors JavaRDD<Vector> data = sc.parallelize(Arrays.asList( Vectors.dense(1.0, 2.0, 3.0), Vectors.dense(4.0, 5.0, 6.0))); Vector transformingVector = Vectors.dense(0.0, 1.0, 2.0); ElementwiseProduct transformer = new ElementwiseProduct(transformingVector); // Batch transform and per-row transform give the same results: JavaRDD<Vector> transformedData = transformer.transform(data); JavaRDD<Vector> transformedData2 = data.map( new Function<Vector, Vector>() { @Override public Vector call(Vector v) { return transformer.transform(v); } });
import org.apache.spark.mllib.regression.LinearRegressionWithSGD import org.apache.spark.mllib.regression.LabeledPoint import org.apache.spark.mllib.linalg.Vectors import org.apache.spark.mllib.feature.PCA val data = sc.textFile("data/mllib/ridge-data/lpsa.data").map { line => val parts = line.split(‘,‘) LabeledPoint(parts(0).toDouble, Vectors.dense(parts(1).split(‘ ‘).map(_.toDouble))) }.cache() val splits = data.randomSplit(Array(0.6, 0.4), seed = 11L) val training = splits(0).cache() val test = splits(1) val pca = new PCA(training.first().features.size/2).fit(data.map(_.features)) val training_pca = training.map(p => p.copy(features = pca.transform(p.features))) val test_pca = test.map(p => p.copy(features = pca.transform(p.features))) val numIterations = 100 val model = LinearRegressionWithSGD.train(training, numIterations) val model_pca = LinearRegressionWithSGD.train(training_pca, numIterations) val valuesAndPreds = test.map { point => val score = model.predict(point.features) (score, point.label)} val valuesAndPreds_pca = test_pca.map { point => val score = model_pca.predict(point.features) (score, point.label)} val MSE = valuesAndPreds.map{case(v, p) => math.pow((v - p), 2)}.mean() val MSE_pca = valuesAndPreds_pca.map{case(v, p) => math.pow((v - p), 2)}.mean() println("Mean Squared Error = " + MSE)println("PCA Mean Squared Error = " + MSE_pca)
原文:http://www.cnblogs.com/yuguoshuo/p/6265777.html