
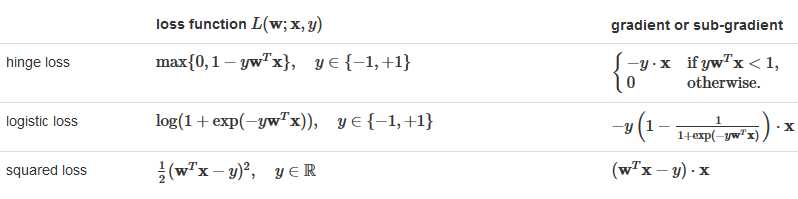
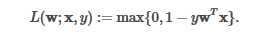
import scala.Tuple2; import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.classification.*; import org.apache.spark.mllib.evaluation.BinaryClassificationMetrics; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.util.MLUtils; import org.apache.spark.SparkConf; import org.apache.spark.SparkContext; public class SVMClassifier { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("SVM Classifier Example"); SparkContext sc = new SparkContext(conf); String path = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint> training = data.sample(false, 0.6, 11L); training.cache(); JavaRDD<LabeledPoint> test = data.subtract(training); // Run training algorithm to build the model. int numIterations = 100; final SVMModel model = SVMWithSGD.train(training.rdd(), numIterations); // Clear the default threshold. model.clearThreshold(); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> scoreAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double score = model.predict(p.features()); return new Tuple2<Object, Object>(score, p.label()); } } ); // Get evaluation metrics. BinaryClassificationMetrics metrics = new BinaryClassificationMetrics(JavaRDD.toRDD(scoreAndLabels)); double auROC = metrics.areaUnderROC(); System.out.println("Area under ROC = " + auROC); // Save and load model model.save(sc, "myModelPath"); SVMModel sameModel = SVMModel.load(sc, "myModelPath"); }}
import org.apache.spark.mllib.optimization.L1Updater; SVMWithSGD svmAlg = new SVMWithSGD();svmAlg.optimizer() .setNumIterations(200) .setRegParam(0.1) .setUpdater(new L1Updater());final SVMModel modelL1 = svmAlg.run(training.rdd());
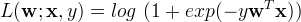
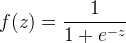
import scala.Tuple2; import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.classification.LogisticRegressionModel; import org.apache.spark.mllib.classification.LogisticRegressionWithLBFGS; import org.apache.spark.mllib.evaluation.MulticlassMetrics; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.util.MLUtils; import org.apache.spark.SparkConf; import org.apache.spark.SparkContext; public class MultinomialLogisticRegressionExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("SVM Classifier Example"); SparkContext sc = new SparkContext(conf); String path = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[] {0.6, 0.4}, 11L); JavaRDD<LabeledPoint> training = splits[0].cache(); JavaRDD<LabeledPoint> test = splits[1]; // Run training algorithm to build the model. final LogisticRegressionModel model = new LogisticRegressionWithLBFGS() .setNumClasses(10) .run(training.rdd()); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> predictionAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double prediction = model.predict(p.features()); return new Tuple2<Object, Object>(prediction, p.label()); } } ); // Get evaluation metrics. MulticlassMetrics metrics = new MulticlassMetrics(predictionAndLabels.rdd()); double precision = metrics.precision(); System.out.println("Precision = " + precision); // Save and load model model.save(sc, "myModelPath"); LogisticRegressionModel sameModel = LogisticRegressionModel.load(sc, "myModelPath"); }}
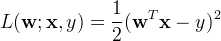
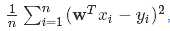
import scala.Tuple2; import org.apache.spark.api.java.*; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.regression.LinearRegressionModel; import org.apache.spark.mllib.regression.LinearRegressionWithSGD; import org.apache.spark.SparkConf; public class LinearRegression { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("Linear Regression Example"); JavaSparkContext sc = new JavaSparkContext(conf); // Load and parse the data String path = "data/mllib/ridge-data/lpsa.data"; JavaRDD<String> data = sc.textFile(path); JavaRDD<LabeledPoint> parsedData = data.map( new Function<String, LabeledPoint>() { public LabeledPoint call(String line) { String[] parts = line.split(","); String[] features = parts[1].split(" "); double[] v = new double[features.length]; for (int i = 0; i < features.length - 1; i++) v[i] = Double.parseDouble(features[i]); return new LabeledPoint(Double.parseDouble(parts[0]), Vectors.dense(v)); } } ); parsedData.cache(); // Building the model int numIterations = 100; final LinearRegressionModel model = LinearRegressionWithSGD.train(JavaRDD.toRDD(parsedData), numIterations); // Evaluate model on training examples and compute training error JavaRDD<Tuple2<Double, Double>> valuesAndPreds = parsedData.map( new Function<LabeledPoint, Tuple2<Double, Double>>() { public Tuple2<Double, Double> call(LabeledPoint point) { double prediction = model.predict(point.features()); return new Tuple2<Double, Double>(prediction, point.label()); } } ); double MSE = new JavaDoubleRDD(valuesAndPreds.map( new Function<Tuple2<Double, Double>, Object>() { public Object call(Tuple2<Double, Double> pair) { return Math.pow(pair._1() - pair._2(), 2.0); } } ).rdd()).mean(); System.out.println("training Mean Squared Error = " + MSE); // Save and load model model.save(sc.sc(), "myModelPath"); LinearRegressionModel sameModel = LinearRegressionModel.load(sc.sc(), "myModelPath"); }}
原文:http://www.cnblogs.com/yuguoshuo/p/6265741.html