算法工程师不仅要搭建模型,还要对模型进行优化及相关线上部署。这里面涉及到很多方面:特征处理(独热编码、归一化)、自定义损失函数、自定义评价函数、超参调节、构建pipeline流程线上部署(100ms返回要求)等。
对于LR模型,进行独热编码(类别型)和归一化(数值型)是很有必要的。比如某个类别型特征的枚举值是0到9(类别型再输入到模型前都会做label enconding,比如有三个枚举值:A、B、C,通过label enconding都变成0、1、2),某个数值型的分布范围是0-1000,那么输入到LR中,模型会认为这两个特征是同一类,就会给数值型特征一个相对小的权重(比如0.001),而给数值型特征一个相对大的权重,但这显然不是我们想要的。因此需要把类别型和数值型特征都转换成0-1之间的数字。
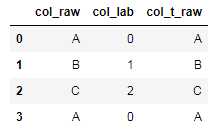
import pandas as pd df_raw=pd.DataFrame([‘A‘,‘B‘,‘C‘,‘A‘],columns=[‘col_raw‘]) #对类别型进行转换 from sklearn import preprocessing lbl = preprocessing.LabelEncoder() col=‘col_raw‘ df_raw[‘col_lab‘] = lbl.fit_transform(df_raw[col].astype(str)) #保存label转换映射关系 import pickle save_lbl_path=‘./onehot_model/‘+col+‘.pkl‘ output = open(save_lbl_path, ‘wb‘) pickle.dump(lbl, output) output.close() #读取和转换 pkl_path=‘./onehot_model/‘+col+‘.pkl‘ pkl_file = open(pkl_path, ‘rb‘) le_departure = pickle.load(pkl_file) df_raw[‘col_t_raw‘] = le_departure.inverse_transform(df_raw[‘col_lab‘])
df_raw
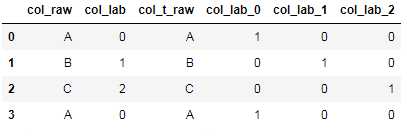
df_cate_tmp=pd.get_dummies(df_raw[‘col_lab‘],prefix=‘col_lab‘) df_raw=pd.concat([df_raw, df_cate_tmp],axis=1) df_raw
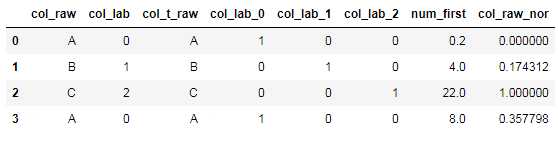
归一化是把数值型特征映射到0-1值域区间,而标准化是把数值分布变成均值为0方差为1标准正态分布。
nor_model=[] col_num=‘num_first‘ #赋值数值型 df_raw[col_num]=[0.2,4,22,8] tmp_min=df_raw[col_num].min() tmp_max=df_raw[col_num].max() nor_model.append((col_num,tmp_min,tmp_max)) #最大-最小归一化法 df_raw[col+‘_nor‘]=df_raw[col_num].apply(lambda x:(x-tmp_min)/(tmp_max-tmp_min)) #保存对应的列名及最大值、最小值 with open("./nor_model/col_min_max.txt","w") as f: for i in nor_model: result=i[0]+‘,‘+str(i[1])+‘,‘+str(i[2])+‘\n‘ f.write(str(result))
在不同的业务场景中,python包提供的损失函数满足不了我们的项目要求,这个时候就需要对自定义损失函数。(参考:https://cloud.tencent.com/developer/article/1357671)
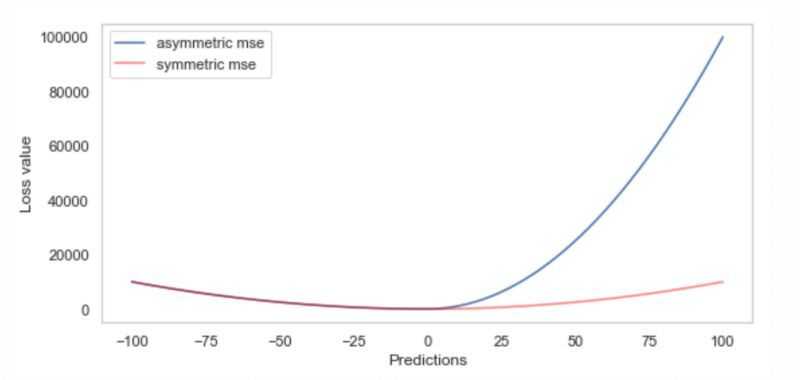
正如定义的那样,非对称MSE很好,因为它很容易计算梯度和hessian,如下图所示。注意,hessian在两个不同的值上是常量,左边是2,右边是20,尽管在下面的图中很难看到这一点。
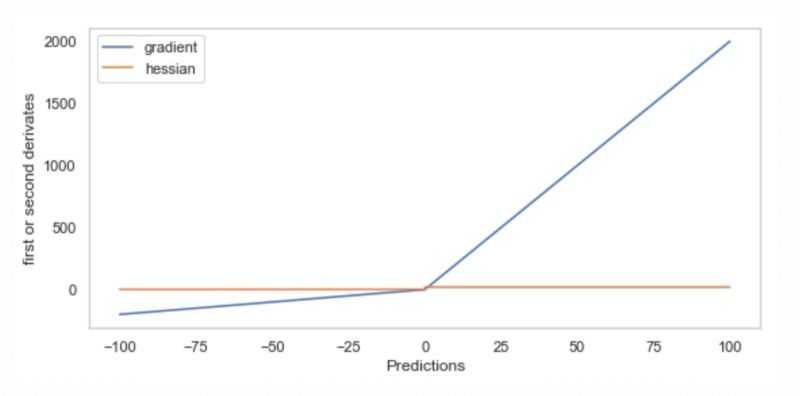
LightGBM提供了一种直接实现定制训练和验证损失的方法。其他的梯度提升包,包括XGBoost和Catboost,也提供了这个选项。这里是一个Jupyter笔记本,展示了如何实现自定义培训和验证损失函数。细节在笔记本上,但在高层次上,实现略有不同。
1、训练损失:在LightGBM中定制训练损失需要定义一个包含两个梯度数组的函数,目标和它们的预测。反过来,该函数应该返回梯度的两个梯度和每个观测值的hessian数组。如上所述,我们需要使用微积分来派生gradient和hessian,然后在Python中实现它。
2、验证丢失:在LightGBM中定制验证丢失需要定义一个函数,该函数接受相同的两个数组,但返回三个值: 要打印的名称为metric的字符串、损失本身以及关于是否更高更好的布尔值。
#---sklearn api def custom_asymmetric_train(y_true, y_pred): residual = (y_true - y_pred).astype("float") grad = np.where(residual<0, -2*10.0*residual, -2*residual) hess = np.where(residual<0, 2*10.0, 2.0) return grad, hess def custom_asymmetric_valid(y_true, y_pred): residual = (y_true - y_pred).astype("float") loss = np.where(residual < 0, (residual**2)*10.0, residual**2) return "custom_asymmetric_eval", np.mean(loss), False #---lgb api def custom_asymmetric_train(preds,dtrain): y_true=np.array(dtrain.get_label()) y_pred=np.argmax(preds.reshape(len(y_true),-1), axis=0) residual = np.array((y_pred-y_true)).astype("float") p=20#参数 tmpGrad=[] tmpHess=[] for i in residual: if i<0: tmpGrad.append(-i*p) tmpHess.append(p) elif (i>=0 and i<=12): tmpGrad.append(i*(p/10)) tmpHess.append(p/10) else: tmpGrad.append(i*p) tmpHess.append(p) grad=np.array(tmpGrad) hess=np.array(tmpHess) return grad, hess def custom_asymmetric_valid(preds,dtrain): p=20#参数 y_true=np.array(dtrain.get_label()) y_pred=np.argmax(preds.reshape(len(y_true),-1), axis=0) residual = np.array((y_pred-y_true)).astype("float") tmpLoss=[] for i in residual: if i<0: tmpLoss.append(-i*p) elif (i>=0 and i<=12): tmpLoss.append(i*(p/10)) else: tmpLoss.append(i*p) loss=np.array(tmpLoss) return "custom_asymmetric_eval", np.mean(loss), False
相应的调用代码(这里需要区分lgb和sklearn的lgb,两个模块的y_red、y是不同的格式):
import lightgbm ********* Sklearn API ********** # default lightgbm model with sklearn api gbm = lightgbm.LGBMRegressor() # updating objective function to custom # default is "regression" # also adding metrics to check different scores gbm.set_params(**{‘objective‘: custom_asymmetric_train}, metrics = ["mse", ‘mae‘]) # fitting model gbm.fit( X_train, y_train, eval_set=[(X_valid, y_valid)], eval_metric=custom_asymmetric_valid, verbose=False, ) y_pred = gbm.predict(X_valid) ********* Python API ********** # create dataset for lightgbm # if you want to re-use data, remember to set free_raw_data=False lgb_train = lgb.Dataset(X_train, y_train, free_raw_data=False) lgb_eval = lgb.Dataset(X_valid, y_valid, reference=lgb_train, free_raw_data=False) # specify your configurations as a dict params = { ‘objective‘: ‘regression‘, ‘verbose‘: 0 } gbm = lgb.train(params, lgb_train, num_boost_round=10, init_model=gbm, fobj=custom_asymmetric_train, feval=custom_asymmetric_valid, valid_sets=lgb_eval) y_pred = gbm.predict(X_valid)
在评估模型预测结果时,如果python库中没有对应的指标(比如k-s值、GINI纯度等),就需要自定义scoring。---此处是针对sklearn的metrics。
from sklearn.metrics import * SCORERS.keys() #结果 dict_keys([‘explained_variance‘, ‘r2‘, ‘neg_median_absolute_error‘, ‘neg_mean_absolute_error‘, ‘neg_mean_squared_error‘, ‘neg_mean_squared_log_error‘, ‘accuracy‘, ‘roc_auc‘, ‘balanced_accuracy‘, ‘average_precision‘, ‘neg_log_loss‘, ‘brier_score_loss‘, ‘adjusted_rand_score‘, ‘homogeneity_score‘, ‘completeness_score‘, ‘v_measure_score‘, ‘mutual_info_score‘, ‘adjusted_mutual_info_score‘, ‘normalized_mutual_info_score‘, ‘fowlkes_mallows_score‘, ‘precision‘, ‘precision_macro‘, ‘precision_micro‘, ‘precision_samples‘, ‘precision_weighted‘, ‘recall‘, ‘recall_macro‘, ‘recall_micro‘, ‘recall_samples‘, ‘recall_weighted‘, ‘f1‘, ‘f1_macro‘, ‘f1_micro‘, ‘f1_samples‘, ‘f1_weighted‘])
import numpy as np def ginic(actual, pred): actual = np.asarray(actual) # In case, someone passes Series or list n = len(actual) a_s = actual[np.argsort(pred)] a_c = a_s.cumsum() giniSum = a_c.sum() / a_s.sum() - (n + 1) / 2.0 return giniSum / n def gini_normalizedc(a, p): if p.ndim == 2: # Required for sklearn wrapper p = p[:,1] # If proba array contains proba for both 0 and 1 classes, just pick class 1 return ginic(a, p) / ginic(a, a) from sklearn import metrics gini_sklearn = metrics.make_scorer(gini_normalizedc, True, True) import numpy as np import pandas as pd from scipy.stats import ks_2samp from sklearn.metrics import make_scorer, roc_auc_score, log_loss,f1_score from sklearn.model_selection import GridSearchCV def ks_stat(y, yhat): y=np.array(y) yhat=np.array(yhat) try: ks =ks_2samp(yhat[y==1],yhat[y!=1]).statistic except: print(yhat.shape,y.shape) kmp=yhat[y==1] kmp2=yhat[y!=1] print(kmp.shape,kmp2.shape) ks=0 return ks ks_scorer = make_scorer(ks_stat, needs_proba=True, greater_is_better=True) log_scorer = make_scorer(log_loss, needs_proba=True, greater_is_better=False) roc_scorer = make_scorer(roc_auc_score, needs_proba=True) f1_scorer = make_scorer(f1_score, needs_proba=True, greater_is_better=True)
模型有很多超参数,通过格点搜索进行(此处,如果设置并行n_jobs>1,则需要把自定义的scoring放在一个外部py文件中,因为:如果在声明函数之前声明池,则尝试并行使用它将引发此错误。颠倒顺序,它将不再抛出此错误)。
from sklearn.ensemble import GradientBoostingClassifier #必须从外部导入scoring import multiprocessing #multiprocessing.set_start_method(‘spawn‘) from external import * from sklearn.model_selection import GridSearchCV tuned_param = {‘learning_rate‘: [0.1, 0.2, 0.5, 0.8, 1], ‘max_depth‘:[3,5,7,10], ‘min_samples_split‘:[50,100,200], ‘n_estimators‘:[50,70,100,150,200], ‘subsample‘:[0.6,0.8]} # n=20 clf_gbdt=GridSearchCV(GradientBoostingClassifier(),tuned_param,cv=2,scoring={‘auc‘: roc_scorer, ‘k-s‘: ks_scorer}, refit=‘k-s‘,n_jobs=n, verbose=10) clf_gbdt.fit(x_train[col_gbdt],y_train) #获取最优超参组合 def print_best_score(gsearch,param_test): # 输出best score print("Best score: %0.3f" % gsearch.best_score_) print("Best parameters set:") # 输出最佳的分类器到底使用了怎样的参数 best_parameters = gsearch.best_estimator_.get_params() for param_name in sorted(param_test.keys()): print("\t%s: %r" % (param_name, best_parameters[param_name])) print_best_score(clf_gbdt,tuned_param)
对于工业界算法模型,模型的稳定是首要!
from sklearn.model_selection import cross_val_score scores=cross_val_score(estimator=lr,X=x_train[col_lr],y=y_train,cv=5,n_jobs=10,scoring=ks_scorer,verbose=10) print(‘CV k-s scores: %s‘%scores) print(‘CV k-s: %.3f +/- %.3f‘%(np.mean(scores),np.std(scores)))
from sklearn.ensemble import GradientBoostingClassifier gbdt = GradientBoostingClassifier(learning_rate=0.2,max_depth=10,min_samples_split=200,n_estimators=200, subsample=0.8,min_samples_leaf=50) gbdt.fit(x_train[col_gbdt], y_train) #模型保存 pickle.dump(gbdt, open(‘./ml_model/gbdt_model.pkl‘, ‘wb‘)) #模型预测输出概率值 y_pred_gbdt=gbdt.predict_proba(x_valid[col_gbdt])
import seaborn as sns sns.set_style(‘darkgrid‘) import matplotlib.pyplot as plt plt.rcParams[‘font.sans-serif‘] = [‘SimHei‘] plt.rcParams[‘axes.unicode_minus‘] = False plt.rcParams.update({‘font.size‘: 10}) plt.rcParams[‘savefig.dpi‘] = 300 #图片像素 plt.rcParams[‘figure.dpi‘] = 300 #分辨率 # 计算AUC fpr_lr,tpr_lr,thresholds = roc_curve(y_valid,y_pred_lr[:,1],pos_label=1) roc_auc_lr = auc(fpr_lr, tpr_lr) # 绘制roc plt.rcParams[‘figure.figsize‘]=(8,5) plt.figure() plt.plot(fpr_lr, tpr_lr, color=‘darkorange‘, label=‘ROC curve (area = %0.2f)‘ % roc_auc_lr) plt.plot([0, 1], [0, 1], color=‘navy‘, linestyle=‘--‘) plt.xlim([0.0, 1.0]) plt.ylim([0.0, 1.05]) plt.xlabel(‘False Positive Rate‘) plt.ylabel(‘True Positive Rate‘) plt.title(‘ROC曲线-LR‘) plt.legend(loc="lower right")
# 绘制K-S曲线 import numpy as np import pandas as pd def PlotKS(preds, labels, n, asc): # preds is score: asc=1 # preds is prob: asc=0 pred = preds # 预测值 bad = labels # 取1为bad, 0为good ksds = pd.DataFrame({‘bad‘: bad, ‘pred‘: pred}) ksds[‘good‘] = 1 - ksds.bad if asc == 1: ksds1 = ksds.sort_values(by=[‘pred‘, ‘bad‘], ascending=[True, True]) elif asc == 0: ksds1 = ksds.sort_values(by=[‘pred‘, ‘bad‘], ascending=[False, True]) ksds1.index = range(len(ksds1.pred)) ksds1[‘cumsum_good1‘] = 1.0*ksds1.good.cumsum()/sum(ksds1.good) ksds1[‘cumsum_bad1‘] = 1.0*ksds1.bad.cumsum()/sum(ksds1.bad) if asc == 1: ksds2 = ksds.sort_values(by=[‘pred‘, ‘bad‘], ascending=[True, False]) elif asc == 0: ksds2 = ksds.sort_values(by=[‘pred‘, ‘bad‘], ascending=[False, False]) ksds2.index = range(len(ksds2.pred)) ksds2[‘cumsum_good2‘] = 1.0*ksds2.good.cumsum()/sum(ksds2.good) ksds2[‘cumsum_bad2‘] = 1.0*ksds2.bad.cumsum()/sum(ksds2.bad) # ksds1 ksds2 -> average ksds = ksds1[[‘cumsum_good1‘, ‘cumsum_bad1‘]] ksds[‘cumsum_good2‘] = ksds2[‘cumsum_good2‘] ksds[‘cumsum_bad2‘] = ksds2[‘cumsum_bad2‘] ksds[‘cumsum_good‘] = (ksds[‘cumsum_good1‘] + ksds[‘cumsum_good2‘])/2 ksds[‘cumsum_bad‘] = (ksds[‘cumsum_bad1‘] + ksds[‘cumsum_bad2‘])/2 # ks ksds[‘ks‘] = ksds[‘cumsum_bad‘] - ksds[‘cumsum_good‘] ksds[‘tile0‘] = range(1, len(ksds.ks) + 1) ksds[‘tile‘] = 1.0*ksds[‘tile0‘]/len(ksds[‘tile0‘]) qe = list(np.arange(0, 1, 1.0/n)) qe.append(1) qe = qe[1:] ks_index = pd.Series(ksds.index) ks_index = ks_index.quantile(q = qe) ks_index = np.ceil(ks_index).astype(int) ks_index = list(ks_index) ksds = ksds.loc[ks_index] ksds = ksds[[‘tile‘, ‘cumsum_good‘, ‘cumsum_bad‘, ‘ks‘]] ksds0 = np.array([[0, 0, 0, 0]]) ksds = np.concatenate([ksds0, ksds], axis=0) ksds = pd.DataFrame(ksds, columns=[‘tile‘, ‘cumsum_good‘, ‘cumsum_bad‘, ‘ks‘]) ks_value = ksds.ks.max() ks_pop = ksds.tile[ksds.ks.idxmax()] tmp_str=‘ks_value is ‘ + str(np.round(ks_value, 4)) + ‘ at pop = ‘ + str(np.round(ks_pop, 4)) # chart # chart plt.plot(ksds.tile, ksds.cumsum_good, label=‘cum_good‘, color=‘blue‘, linestyle=‘-‘, linewidth=2) plt.plot(ksds.tile, ksds.cumsum_bad, label=‘cum_bad‘, color=‘red‘, linestyle=‘-‘, linewidth=2) plt.plot(ksds.tile, ksds.ks, label=‘ks‘, color=‘green‘, linestyle=‘-‘, linewidth=2) plt.axvline(ks_pop, color=‘gray‘, linestyle=‘--‘) plt.axhline(ks_value, color=‘green‘, linestyle=‘--‘) plt.axhline(ksds.loc[ksds.ks.idxmax(), ‘cumsum_good‘], color=‘blue‘, linestyle=‘--‘) plt.axhline(ksds.loc[ksds.ks.idxmax(),‘cumsum_bad‘], color=‘red‘, linestyle=‘--‘) plt.title(‘KS=%s ‘ %np.round(ks_value, 4) + ‘at Pop=%s‘ %np.round(ks_pop, 4), fontsize=15) return tmp_str
#调用
PlotKS(y_valid,y_pred_lr[:,1], n=10, asc=0)
#根据上述可知,在0.2的时候区分度最好,因此认为大于0.2的就是1,小于则为0 y_pred_lr_new=[] for i in y_pred_lr[:,1]: if i<=0.2: y_pred_lr_new.append(0) else: y_pred_lr_new.append(1) y_pred_gbdt_new=[] for i in y_pred_gbdt[:,1]: if i<=0.2: y_pred_gbdt_new.append(0) else: y_pred_gbdt_new.append(1) y_pred_lr_gbdt_new=[] for i in y_pred_gbdt_lr[:,1]: if i<=0.2: y_pred_lr_gbdt_new.append(0) else: y_pred_lr_gbdt_new.append(1) # gbdt acc=accuracy_score(y_valid, y_pred_gbdt_new) p = precision_score(y_valid, y_pred_gbdt_new, average=‘binary‘) r = recall_score(y_valid, y_pred_gbdt_new, average=‘binary‘) f1score = f1_score(y_valid, y_pred_gbdt_new, average=‘binary‘) print(acc,p,r,f1score)
初始模型以读取pickle文件方式:首先通过python脚本读取pickle文件,再起flask提供模型预测服务,最后java应用调用flask实现预测。
用java脚本实现LR模型类:通过java脚本构建模型(通过python训练好的模型可以输出成规则)方法,java应用之间调这个方法实现预测。
使用Java调用jpmml:把python相关的模型(独热编码、LR模型等)转成PMML文件(相当于一个本地txt文件),再通过java类(jpmml)调用。
综上所述,综合考虑计算速度及计算资源,推荐使用方案3。
参考链接:https://openscoring.io/blog/2019/06/19/sklearn_gbdt_lr_ensemble/
from lightgbm import LGBMClassifier from sklearn_pandas import DataFrameMapper from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier from sklearn.linear_model import LogisticRegression from sklearn.preprocessing import LabelBinarizer, LabelEncoder from sklearn2pmml import sklearn2pmml from sklearn2pmml.decoration import CategoricalDomain, ContinuousDomain from sklearn2pmml.ensemble import GBDTLRClassifier from sklearn2pmml.pipeline import PMMLPipeline from xgboost import XGBClassifier import pandas df = pandas.read_csv("audit.csv") cat_columns = ["Education", "Employment", "Marital", "Occupation"] cont_columns = ["Age", "Hours", "Income"] label_column = "Adjusted" def make_fit_gbdtlr(gbdt, lr): mapper = DataFrameMapper( [([cat_column], [CategoricalDomain(), LabelBinarizer()]) for cat_column in cat_columns] + [(cont_columns, ContinuousDomain())] ) classifier = GBDTLRClassifier(gbdt, lr) pipeline = PMMLPipeline([ ("mapper", mapper), ("classifier", classifier) ]) pipeline.fit(df[cat_columns + cont_columns], df[label_column]) return pipeline pipeline = make_fit_gbdtlr(GradientBoostingClassifier(n_estimators = 499, max_depth = 2), LogisticRegression()) sklearn2pmml(pipeline, "GBDT+LR.pmml") pipeline = make_fit_gbdtlr(RandomForestClassifier(n_estimators = 31, max_depth = 6), LogisticRegression()) sklearn2pmml(pipeline, "RF+LR.pmml") pipeline = make_fit_gbdtlr(XGBClassifier(n_estimators = 299, max_depth = 3), LogisticRegression()) sklearn2pmml(pipeline, "XGB+LR.pmml") def make_fit_lgbmlr(gbdt, lr): mapper = DataFrameMapper( [([cat_column], [CategoricalDomain(), LabelEncoder()]) for cat_column in cat_columns] + [(cont_columns, ContinuousDomain())] ) classifier = GBDTLRClassifier(gbdt, lr) pipeline = PMMLPipeline([ ("mapper", mapper), ("classifier", classifier) ]) pipeline.fit(df[cat_columns + cont_columns], df[label_column], classifier__gbdt__categorical_feature = range(0, len(cat_columns))) return pipeline pipeline = make_fit_lgbmlr(LGBMClassifier(n_estimators = 71, max_depth = 5), LogisticRegression()) sklearn2pmml(pipeline, "LGBM+LR.pmml")
注意:jdk必须是1.8及以上、sklearn的版本必须0.21.0及以上、sklearn2pmml版本最好通过git获取安装
import sklearn import sklearn2pmml print(‘The sklearn2pmml version is {}.‘.format(sklearn2pmml.__version__)) print(‘The scikit-learn version is {}.‘.format(sklearn.__version__)) #多个jdk版本设置切换 export JAVA_HOME=/software/servers/jdk1.8.0_121/ export PATH=$JAVA_HOME/bin:$PATH export CLASSPATH=.:$JAVA_HOME/lib/dt.jar:$JAVA_HOME/lib/tools.jar
原文:https://www.cnblogs.com/siriJR/p/12180019.html