目前主流的基因填充方法有两种:一步法填充和两步法填充,其对比如下图
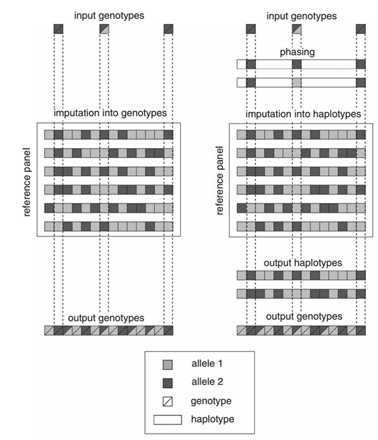
根据参考面板的基因型推断样本可能的基因型构成, 然后直接填充缺失的基因型,这种样本单倍型是根据参考样本的单倍型来 进行推断的,每一个样本都需要推断一次,并且参考样本更改以后,也需要重新根据参考样本来进行单倍型推断。
./impute2 -m ./Example/example.chr22.map -h ./Example/example.chr22.1kG.haps -l ./Example/example.chr22.1kG.legend -g ./Example/example.chr22.study.gens -strand_g ./Example/example.chr22.study.strand -int 20.4e6 20.5e6 -Ne 20000 -o ./Example/example.chr22.one.phased.impute2
其中:
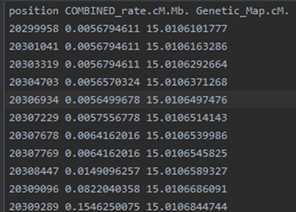
example.chr22.map:
example.chr22.1kG.haps:

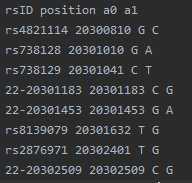
example.chr22.1kG.legend:
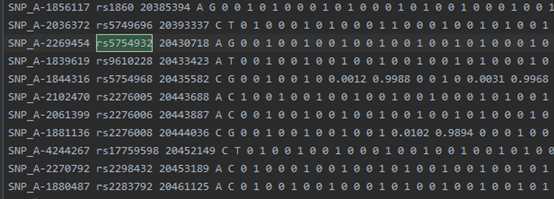
example.chr22.study.gens:
example.chr22.study.strand:
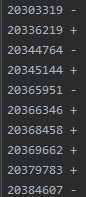
example.chr22.one.phased.impute2:
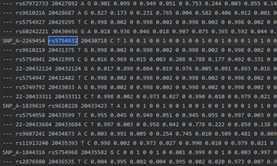
基因分型,是按照亲本正确地定位到父亲或者母亲的染色体上,最终使得所有来自同一个亲本的等位基因都能够排列在同一个染色体里面,基因分型有三种方法:家系分型(Related individuals Phasing)、群体LD分型(LD Phasing)和物理分型(Physical Phasing),其中群体LD和家系分型常用,SHAPEIT2 为比较常用的分型软件。
impute2的原理是通过滑窗的形式进行学习参考分布,然后实时对填充序列进行填充,其实本质上如果这种方式填充序列样本量比较小的时候就跟第二种是一样的,序列的分布情况主要就依赖于参考序列,这种方式类似于进行比对,利用神经网络学习比对的模式,然后进行运用
Step 1: Pre-phasing
./impute2 -prephase_g -m ./Example/example.chr22.map -g ./Example/example.chr22.study.gens -int 20.4e6 20.5e6 -Ne 20000 -o ./Example/example.chr22.prephasing.impute2
Example/example.chr22.map:
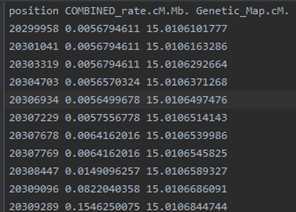
example.chr22.study.gens:
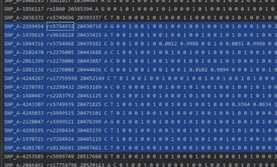
example.chr22.prephasing.impute2:
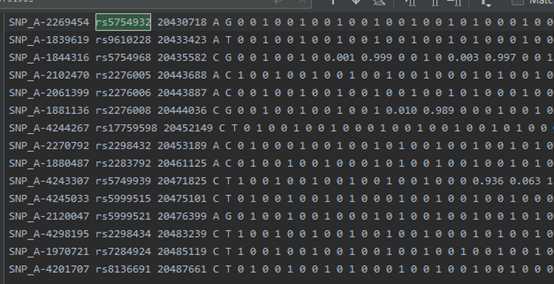
Step 2: Imputation into pre-phased haplotypes
./impute2 -use_prephased_g -m ./Example/example.chr22.map -h ./Example/example.chr22.1kG.haps -l ./Example/example.chr22.1kG.legend -known_haps_g ./Example/example.chr22.prephasing.impute2_haps -strand_g ./Example/example.chr22.study.strand -int 20.4e6 20.5e6 -Ne 20000 -o ./Example/example.chr22.one.phased.impute2
-phase
example.chr22.1kG.legend:
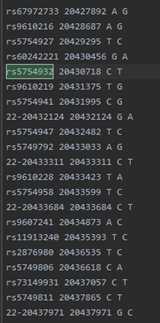
example.chr22.prephasing.impute2_haps:
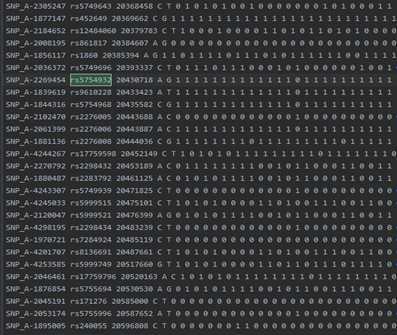
此文件的snp和study的snp数量是一致的。
example.chr22.one.phased.impute2:
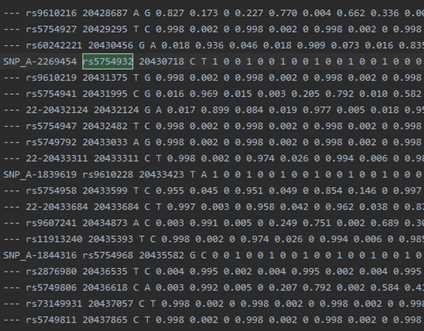
IMPUTATION WITH ONE UNPHASED REFERENCE PANEL
IMPUTATION WITH TWO PHASED REFERENCE PANELS
IMPUTATION WITH TWO PHASED REFERENCE PANELS (MERGE REFERENCE PANELS)
IMPUTATION WITH ONE PHASED AND ONE UNPHASED REFERENCE PANEL
IMPUTATION WITH ONE PHASED AND ONE UNPHASED REFERENCE PANEL, WITH ADDITIONAL OPTIONS
原文:https://www.cnblogs.com/univeryinli/p/13202817.html