求解的问题,burma.tsp里面的内容
1 16.47 96.10 2 16.47 94.44 3 20.09 92.54 4 22.39 93.37 5 25.23 97.24 6 22.00 96.05 7 20.47 97.02 8 17.20 96.29 9 16.30 97.38 10 14.05 98.12 11 16.53 97.38 12 21.52 95.59 13 19.41 97.13 14 20.09 94.55
主程序
为方便调用,已经将所有的模块整合到一个程序内。
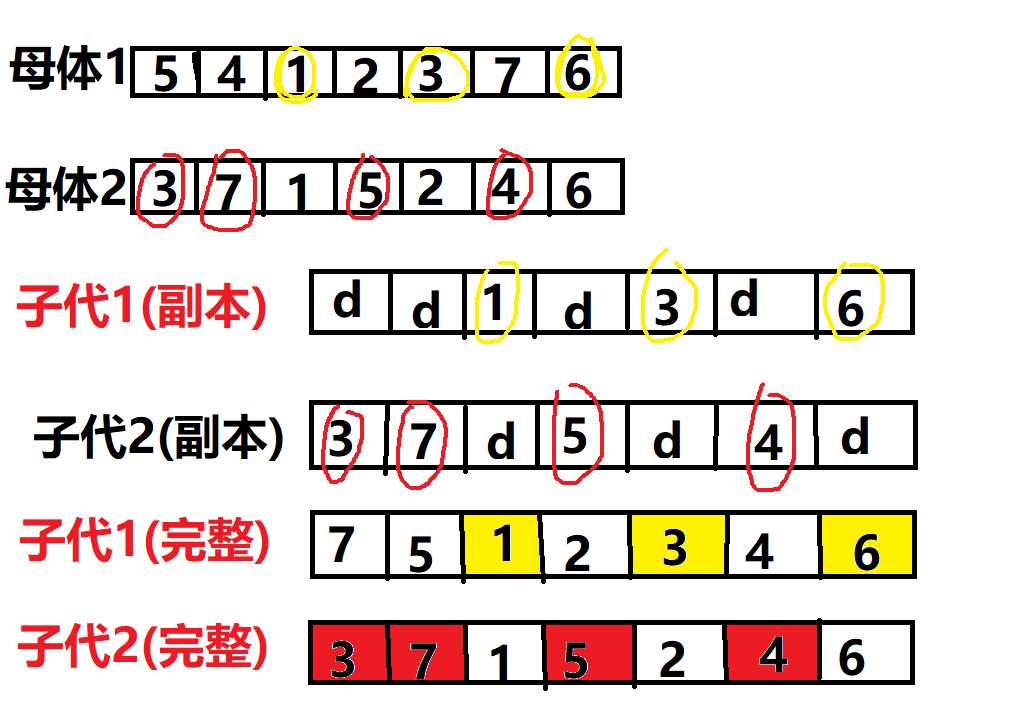
import random import math import matplotlib.pyplot as plt import re #-------参数部分--------# generation=[] fitvalue=[] # dict={ # 1:[16.47,96.10],2:[16.47,94.44],3:[20.09,92.54], # 4:[22.39,93.37],5:[25.23,97.24],6:[22.00,96.05], # 7:[20.47,97.02],8:[17.20,96.29],9:[16.30,97.38], # 10:[14.05,98.12],11:[16.53,97.38],12:[21.52,95.59], # 13:[19.41,97.13],14:[20.09,94.55] # } #修改部位1。此部分的字典操作应该采用文件读写的方式来完成 crossrate=4 mutationrate=1 size=30 popgeneration=400 #——————————————这里是交叉的模块——————————# def crossOver(population): ret = [] for i in range(0,int(len(population)/2)): cplist1=[] cplist2=[] register1=[] register2=[] crossval1=[] crossval2=[] x1=random.choice(population) target=0 while target!=1: x2=random.choice(population) if x2!=x1: target=1 if x2==x1: continue # print(‘随机抽出两个不同的母体‘,(x1,x2)) while len(cplist1)<=crossrate: #随机抽crossrate条染色体位置传给子代 crossposition=random.randrange(0,13) if crossposition not in cplist1: #保证随机取得交叉位置不重复 cplist1.append(crossposition) crossval1.append(x1[crossposition]) # print(‘母体1传给子代的交叉位置%s,交叉位置值%s‘ %(cplist1,crossval1)) for i in range(0,14): if i not in cplist1: cplist2.append(i) crossval2.append(x2[i]) # print(‘母体2传给子代的交叉位置%s,交叉位置值%s‘ %(cplist2,crossval2)) for i1 in range(0,14): if i1 not in cplist1: #单体长度14,如果是交叉位置上的就传值,如果不是就传d register1.append(‘d‘) if i1 in cplist1: register1.append(x1[i1]) # print(‘母体1交叉位置形成子代1副本‘,register1) for i2 in x2: if i2 not in crossval1: #给d更换的值不能与已经存在的值重复 for xp in range(0,len(register1)): if register1[xp]==‘d‘: #用于定位d的位置 register1[xp]=i2 break # print(‘交叉后的子体1‘,register1) for p1 in range(0,14): if p1 not in cplist2: register2.append(‘d‘) if p1 in cplist2: register2.append(x2[p1]) # print(‘母体2交叉位置形成子代1副本‘,register2) for p2 in x1: if p2 not in crossval2: for xp1 in range(0,len(register2)): if register2[xp1]==‘d‘: register2[xp1]=p2 break # print(‘交叉后的子体2‘,register2) ret.append(register1) ret.append(register2) print(‘交叉完成后的种群‘,ret) return ret #——————————————这里是变异的模块——————————# def mutation(population): ret=[] for i in population: porbility=random.uniform(0,1) if porbility<mutationrate: a=random.randrange(0,14) b=random.randrange(0,14) trans=i[a] trans1=i[b] i[a]=trans1 i[b]=trans ret.append(i) else: ret.append(i) # print(‘变异完成后的种群‘,ret) return ret #——————————————————这里是产生随机种群的模块——————————# def creatPop(popsize): ret1=[] for i in range(0,popsize): ret=[] # while(len(ret)<=13): # x=random.randrange(1,15) # if x not in ret: #xulie # ret.append(x) # ret1.append(ret) #根据上次说的改进,可以使用random乱序来生成 for i1 in range(0,14): ret.append(i1+1) random.shuffle(ret) ret1.append(ret) print(‘生成的初代种群‘,ret1) return ret1 creatPop(2) #——————————————————这里是将文件内的数据读取出来并放入字典的程序——————————# dict={} fil=open(‘burma14.tsp‘,‘r‘,encoding=‘utf8‘) #r代表读操作,且打开时按utf8 for i in range(0,14): readtext=fil.readline() readline=re.split(‘[ ]‘,readtext) for i in range(0,4): readline.remove(‘‘) dict.update({int(readline[0]):[float(readline[1]),float(readline[2])]}) print(dict) #————————————————这里是适应度函数值的计算模块—————— def fitNess(population): ret=[] for i in population: # print(i) distance = 0 s=0 for xp in range(0,len(i)-1): # print(dict[i[s]]) # print(dict[i[s+1]]) distance=distance+((dict[i[s]][0]-dict[i[s+1]][0])**2+(dict[i[s]][1]-dict[i[s+1]][1])**2)**(1/2) # print(distance) #每两个相邻的点位求距离 s=s+1 distance=distance+((dict[i[0]][0]-dict[i[13]][0])**2+(dict[i[0]][1]-dict[i[13]][1])**2)**(1/2) #最后再求终点到起点的距离 ret.append(distance) print(‘计算并集内个体适应度‘,ret) return ret def popChoice(population): ret=[] ret1=[] dict1={} unionpop=[] for i in start: unionpop.append(i) for i1 in population: unionpop.append(i1) print(‘新老种群并集‘,unionpop) fitlist=fitNess(unionpop) for xp in range(0,len(fitlist)): dict1.update({fitlist[xp]:unionpop[xp]}) print(dict1) fitlist.sort() print(fitlist) for xp1 in fitlist: ret.append(dict1[xp1]) print(ret) for xp2 in range(0,size): if ret[xp2] not in ret1: ret1.append(ret[xp2]) print(ret1) print(‘最优适应度值为‘,fitlist[0]) fitvalue.append(fitlist[0]) return ret1 a=creatPop(size) start=a for i in range(0,popgeneration): b=crossOver(start) c=mutation(b) d=popChoice(c) print(‘第%s世代,他的最优解为%s‘ %(i+1,d[0])) start=d for i1 in range(0,popgeneration): generation.append(i1) plt.plot(generation,fitvalue) plt.ylabel(‘fitness value‘) #为y轴加注释 plt.xlabel(‘generation‘) #为x轴加注释 plt.show()
原文:https://www.cnblogs.com/zqh962903415/p/13931738.html