DAY3
对于给定的节点,DeepWalk会等概率的选取下一个相邻节点加入路径,直至达到最大路径长度,或者没有下一个节点可选。
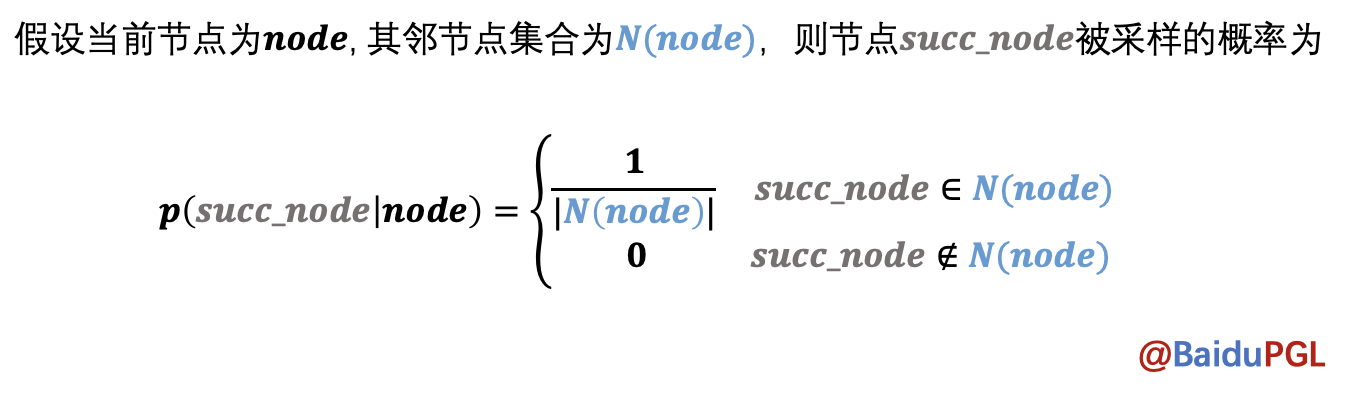

实现代码:
%%writefile userdef_graph.py from pgl.graph import Graph import numpy as np class UserDefGraph(Graph): def random_walk(self, nodes, walk_len): """ 输入:nodes - 当前节点id list (batch_size,) walk_len - 最大路径长度 int 输出:以当前节点为起点得到的路径 list (batch_size, walk_len) 用到的函数 1. self.successor(nodes) 描述:获取当前节点的下一个相邻节点id列表 输入:nodes - list (batch_size,) 输出:succ_nodes - list of list ((num_successors_i,) for i in range(batch_size)) 2. self.outdegree(nodes) 描述:获取当前节点的出度 输入:nodes - list (batch_size,) 输出:out_degrees - list (batch_size,) """ walks = [[node] for node in nodes] walks_ids = np.arange(0, len(nodes)) cur_nodes = np.array(nodes) for l in range(walk_len): """选取有下一个节点的路径继续采样,否则结束""" outdegree = self.outdegree(cur_nodes) walk_mask = (outdegree != 0) if not np.any(walk_mask): break cur_nodes = cur_nodes[walk_mask] walks_ids = walks_ids[walk_mask] outdegree = outdegree[walk_mask] ###################################### # 请在此补充代码采样出下一个节点 succ_nodes = self.successor(cur_nodes) next_nodes = [ np.random.choice(node) for node in succ_nodes] for i, next_node in zip(walks_ids,next_nodes): walks[i].append(next_node) ###################################### cur_nodes = np.array(next_nodes) return walks
NOTE:在得到节点路径后,node2vec会使用SkipGram模型学习节点表示,给定中心节点,预测局部路径中还有哪些节点。模型中用了negative sampling来降低计算量。
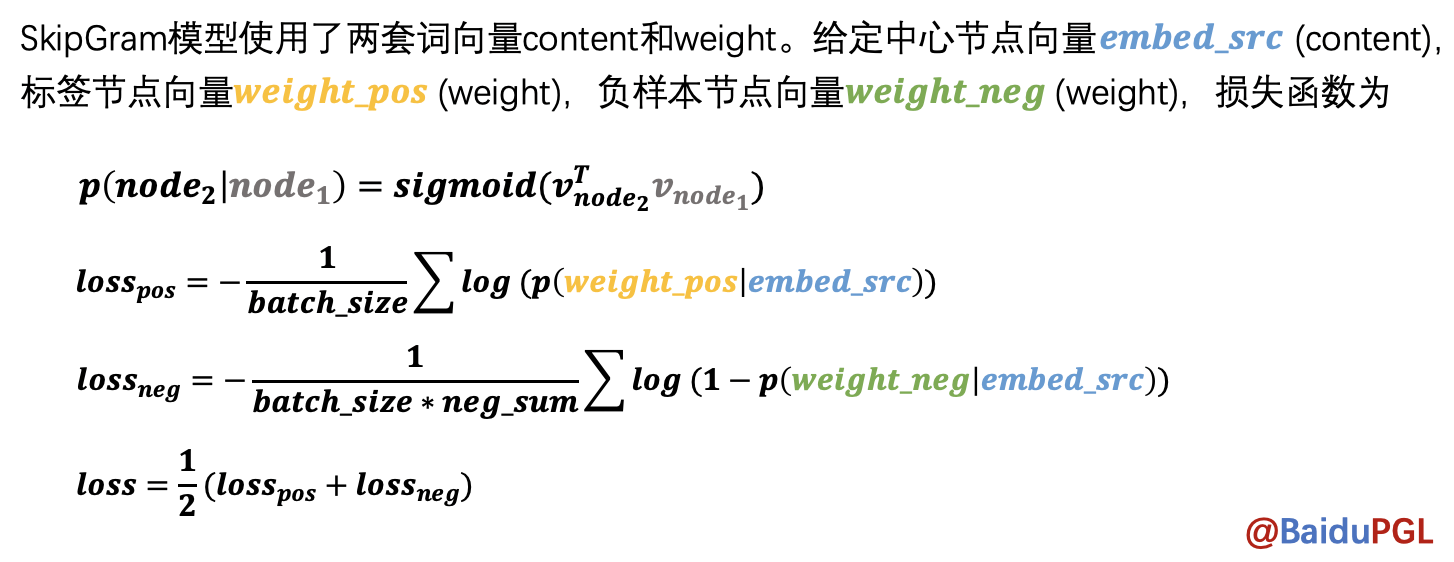

实现代码:
%%writefile userdef_model.py import paddle.fluid.layers as l def userdef_loss(embed_src, weight_pos, weight_negs): """ 输入:embed_src - 中心节点向量 list (batch_size, 1, embed_size) weight_pos - 标签节点向量 list (batch_size, 1, embed_size) weight_negs - 负样本节点向量 list (batch_size, neg_num, embed_size) 输出:loss - 正负样本的交叉熵 float """ """ pos_logits = l.matmul( embed_src, weight_pos, transpose_y=True) # [batch_size, 1, 1] neg_logits = l.matmul( embed_src, weight_negs, transpose_y=True) # [batch_size, 1, neg_num] """ ################################## # 请在这里实现SkipGram的loss计算过程 pos_logits = l.matmul( embed_src, weight_pos, transpose_y=True) # [batch_size, 1, 1] neg_logits = l.matmul( embed_src, weight_negs, transpose_y=True) # [batch_size, 1, neg_num] ones_label = pos_logits * 0. + 1. ones_label.stop_gradient = True pos_loss = l.sigmoid_cross_entropy_with_logits(pos_logits, ones_label) zeros_label = neg_logits * 0. zeros_label.stop_gradient = True neg_loss = l.sigmoid_cross_entropy_with_logits(neg_logits, zeros_label) loss = (l.reduce_mean(pos_loss) + l.reduce_mean(neg_loss)) / 2 ################################## return loss
Node2Vec会根据与上个节点的距离按不同概率采样得到当前节点的下一个节点。
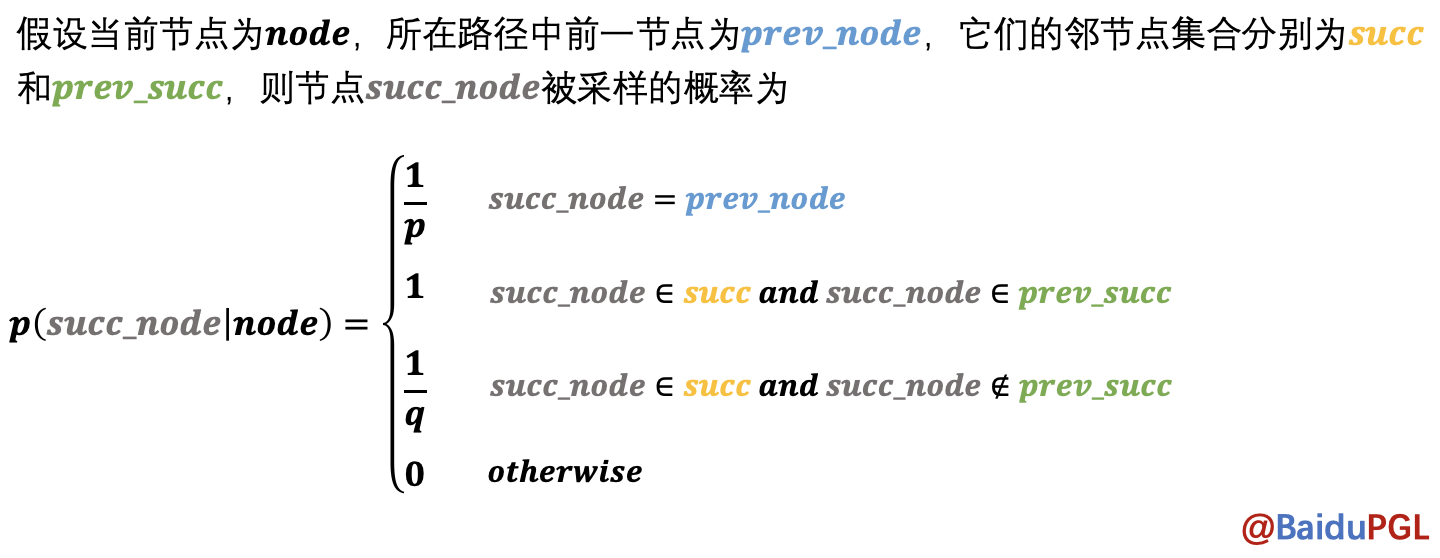
实现代码:
%%writefile userdef_sample.py import numpy as np def node2vec_sample(succ, prev_succ, prev_node, p, q): """ 输入:succ - 当前节点的下一个相邻节点id列表 list (num_neighbors,) prev_succ - 前一个节点的下一个相邻节点id列表 list (num_neighbors,) prev_node - 前一个节点id int p - 控制回到上一节点的概率 float q - 控制偏向DFS还是BFS float 输出:下一个节点id int """ ################################## # 请在此实现node2vec的节点采样函数 probs = [] prob_sum = 0. for succ_ in succ: if succ_ == prev_node: prob = 1. / p elif succ_ in prev_succ: prob = 1. else: prob = 1.0 / q probs.append(prob) prob_sum += prob random_number = np.random.rand() * prob_sum for i, succ_ in enumerate(succ): random_number -= probs[i] if random_number <= 0: sampled_succ = succ_ break ################################## return sampled_succ

GraphSage主要解决图空间太大不够内存计算的问题,采取了类似mini-batch的方式。
原文:https://www.cnblogs.com/bad-joker/p/14057265.html